Classification of SVD-mapped Datasets¶
Demonstrate the usage of a dataset mapper performing data projection onto singular value components within a cross-validation – for any classifier.
from mvpa2.suite import *
if __debug__:
debug.active += ["REPM"]
#
# load PyMVPA example dataset
#
dataset = load_example_fmri_dataset(literal=True)
#
# preprocessing
#
# do chunkswise linear detrending on dataset
poly_detrend(dataset, polyord=1, chunks_attr='chunks')
# only use 'rest', 'cats' and 'scissors' samples from dataset
dataset = dataset[np.array([ l in ['rest', 'cat', 'scissors']
for l in dataset.targets], dtype='bool')]
# zscore dataset relative to baseline ('rest') mean
zscore(dataset, chunks_attr='chunks', param_est=('targets', ['rest']), dtype='float32')
# remove baseline samples from dataset for final analysis
dataset = dataset[dataset.sa.targets != 'rest']
# Specify the class of a base classifier to be used
Clf = LinearCSVMC
# And create the instance of SVDMapper to be reused
svdmapper = SVDMapper()
Lets create a generator of a ChainMapper which would first perform
SVD and then subselect the desired range of components.
get_SVD_sliced = lambda x: ChainMapper([svdmapper,
StaticFeatureSelection(x)])
Now we can define a list of some classifiers: a simple one and several classifiers with built-in SVD transformation and selection of corresponding SVD subspaces
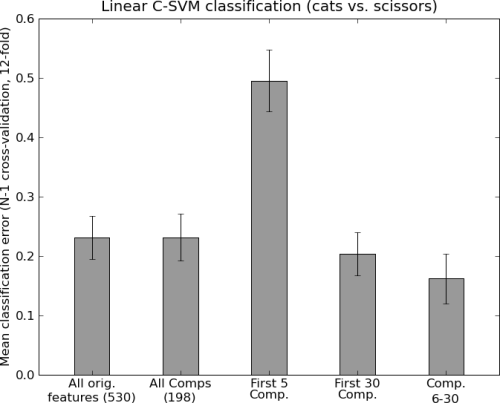
clfs = [('All orig.\nfeatures (%i)' % dataset.nfeatures, Clf()),
('All Comps\n(%i)' % (dataset.nsamples \
- (dataset.nsamples / len(dataset.UC)),),
MappedClassifier(Clf(), svdmapper)),
('First 5\nComp.', MappedClassifier(Clf(),
get_SVD_sliced(slice(0, 5)))),
('First 30\nComp.', MappedClassifier(Clf(),
get_SVD_sliced(slice(0, 30)))),
('Comp.\n6-30', MappedClassifier(Clf(),
get_SVD_sliced(slice(5, 30))))]
# run and visualize in barplot
results = []
labels = []
for desc, clf in clfs:
print desc.replace('\n', ' ')
cv = CrossValidation(clf, NFoldPartitioner())
res = cv(dataset)
# there is only one 'feature' i.e. the error in the returned
# dataset
results.append(res.samples[:,0])
labels.append(desc)
plot_bars(results, labels=labels,
title='Linear C-SVM classification (cats vs. scissors)',
ylabel='Mean classification error (N-1 cross-validation, 12-fold)',
distance=0.5)
Output of the example analysis:
See also
The full source code of this example is included in the PyMVPA source distribution (doc/examples/svdclf.py).




